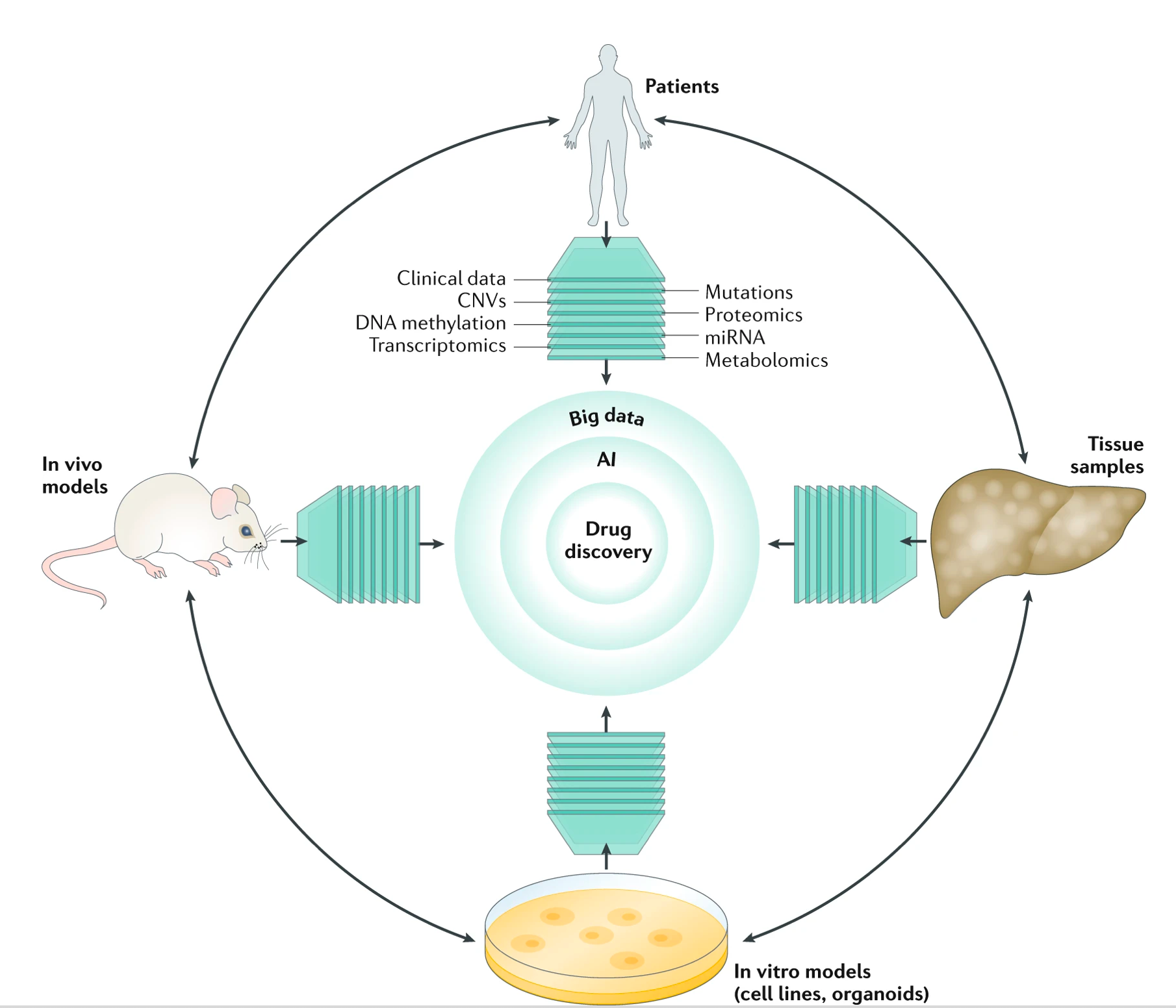
OMICS-based drug discovery
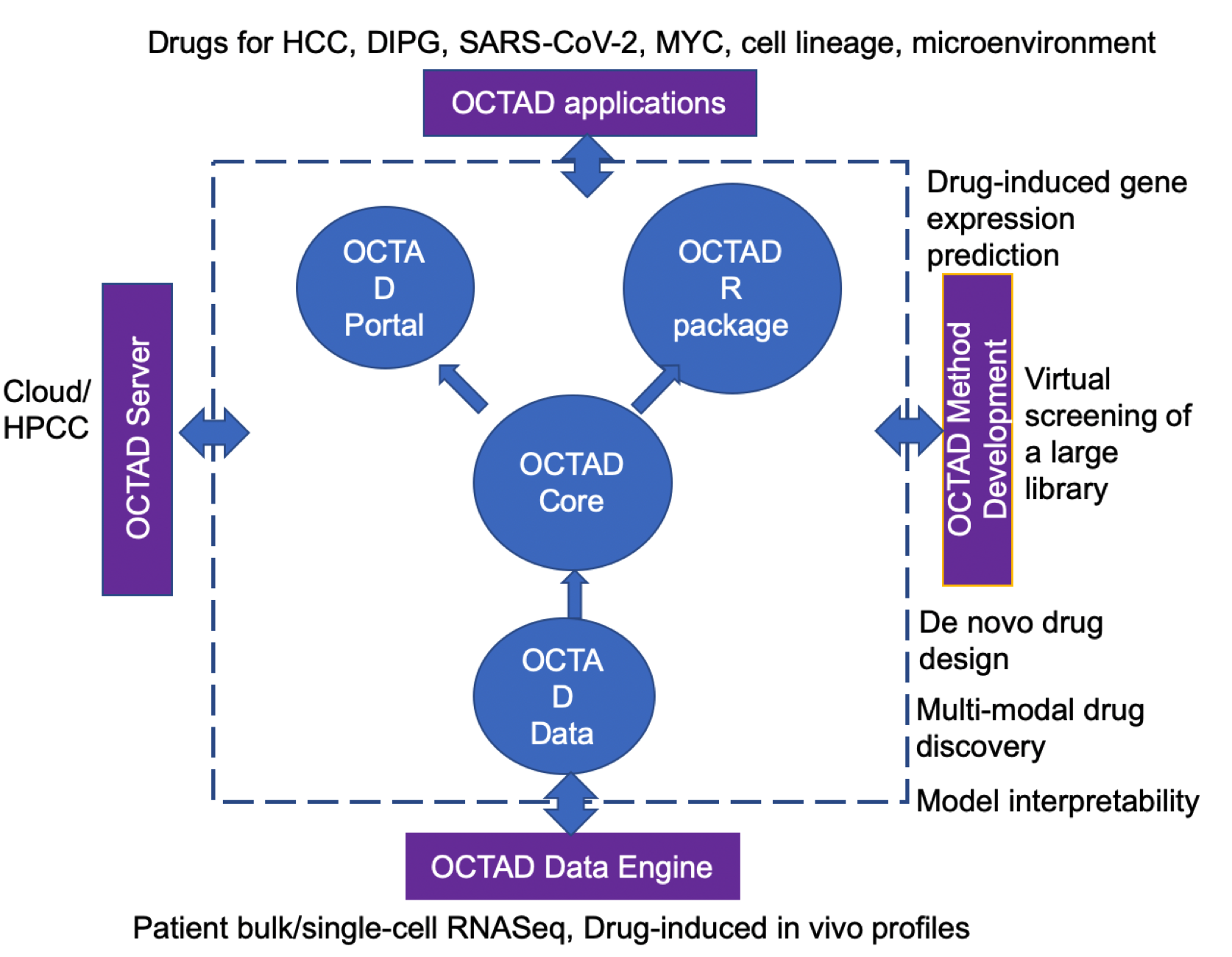
Conventional drug discovery focuses on finding agents that modulate a molecular feature (mostly a protein target). This approach was adopted a few decades ago, even before the emergence of omics technologies. Today’s technologies allow the profiling of thousands of molecular features including proteins, mRNAs, and metabolites, all of which can contribute to disease development. Finding one magic bullet from these thousands of features is challenging. Moreover, focusing only on a single feature does not fully utilize all the information generated from new omics technologies. We employed a systems approach that considers how drugs affect a disease gene expression signature (comprised of a set of up-/down-regulated disease genes). This approach led to the discovery of therapeutic candidates for hepatocellular carcinoma (HCC), Ewing sarcoma, basal cell carcinoma, melanoma, Alzheimer’s disease, DIPG (Diffuse intrinsic pontine glioma), prostate cancer bone metastasis, and SARS-CoV-2. Moreover, we discovered that the potency to reverse gene expression correlates with drug efficacy, providing strong evidence of using this approach in drug repurposing (Chen, Nature Communications, 2017). We developed a drug-repurposing platform called OCTAD (Zeng, Nature Protocols, 2021 ), allowing researchers to predict repurposed drug candidates for cancers and cancer subtypes.
We actively develop new methods to support novel compound screening and lead optimization. Our recent conference papers have demonstrated the feasibility through improving the training data quality (Sun, ICDM, 2020), pre-training graph neural network (Sun, KDD, 2021) and tree search based multi-objective optimization (Sun, KDD, 2022). Now our model can quickly predict novel compounds based on disease gene expression profiles. The lab has set up a bench to validate new candidates for multiple diseases including HCC, diffuse intrinsic pontine glioma (DIPG), and SARS-CoV-2.
With the emergence of single-cell RNASeq data, we plan to apply this approach to identify compounds that could control cell lineage development or modulate the microenvironment. This approach also holds great promise for identifying novel inhibitors for undruggable targets. On the technical side, we will develop methods to support de novo drug design using advanced machine learning.
OMICS-based biomarker and target identification
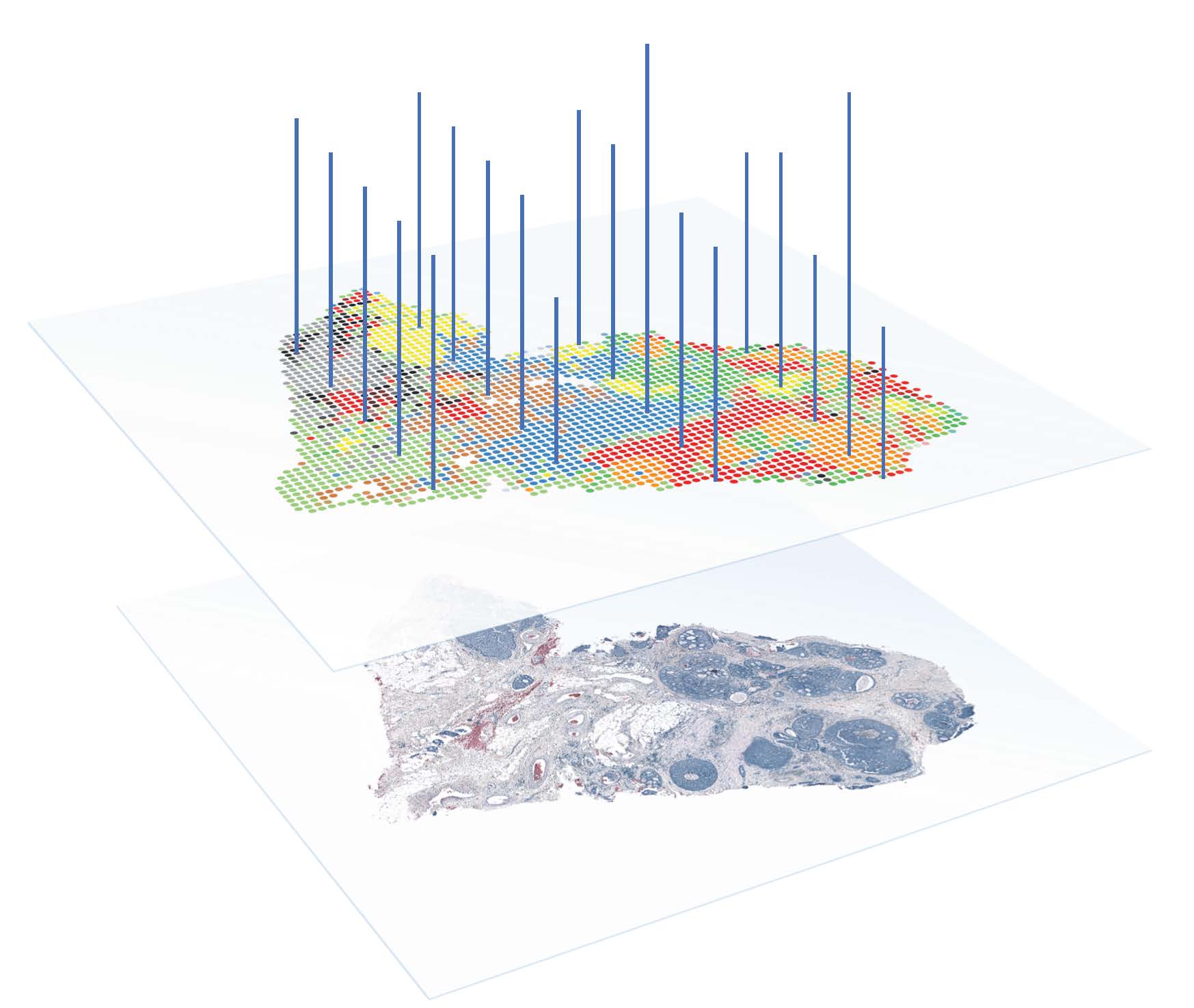
The omics-based drug discovery work could not flourish without an extensive understanding of disease mechanisms. Therefore, the second pillar of the lab research focuses on leveraging emerging single-cell RNASeq, spatial transcriptomics, rich bulk gene expression profiles, and advanced machine learning to identify novel biomarkers and therapeutic targets. We started this endeavor by working with local physicians to predict MODS patients who would need ECMO life support (Shankar, EBioMedicine, 2021). We currently utilize single-cell omics to delineate cell compositions and cell-cell interactions.
In addition to the MODS biomarker work, four projects are undergoing: 1) characterizing microenvironment of liver diseases through in silico characterization of bulk tissues and spatial transcriptomics (GitHub), 2) portraying pan-cancer liver metastasis microenvironment (GitHub), 3) discovering bispecific antibody targets for cancer immunotherapy (GitHub), and 4) predicting surfaceome using single-cell transcriptomics and transfer learning (GitHub).
Virtual platform to support translational drug discovery
In order to increase drug discovery success rates, we have to scrutinize every step in translational research. The lab is dedicated to developing a virtual platform supported by machine learning and data analytics. The platform includes tumor sample selection, reference normal tissue selection, cell line evaluation, pharmacogenomic data correction, and EHR data cleaning and mining ( MetBERT). We recently develop TransCell that allows in-silico characterization of genomic features and cellular responses solely from gene expression data. The lab continues to leverage open data and proprietary data (the EHR data of 2 million patients and >8000 biobank patient specimens at Corewell Health) and develop computational tools to answer interesting questions in translational research.